Options for 3D Cell Culture: The Benefits of PeptiGel

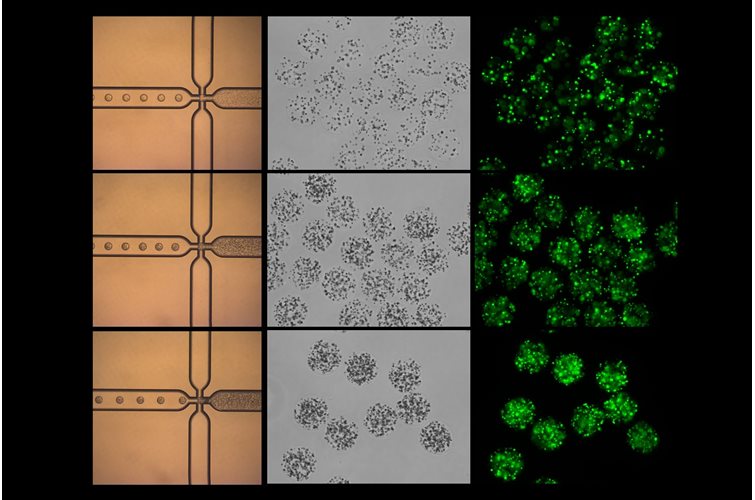
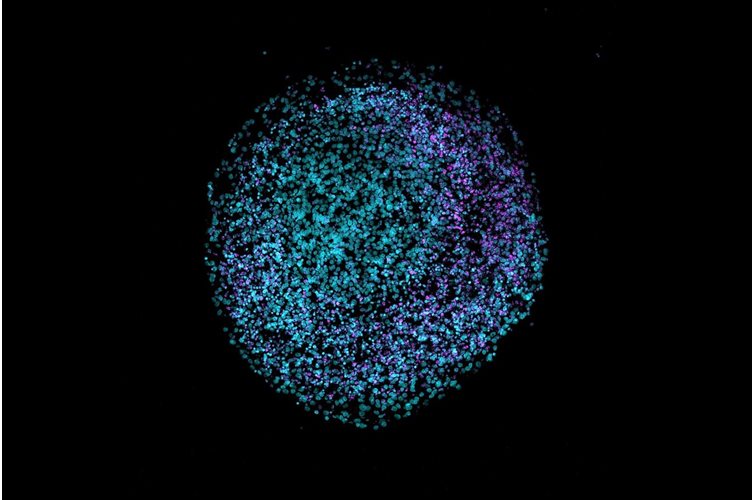
3D cell culture has emerged as a pivotal technique for mimicking the natural environment of cells. This method offers a more physiologically relevant context compared to traditional 2D cultures, better enabling the study of cell behavior, drug responses, and disease mechanisms. In this article, we will delve into the pros and cons of various hydrogels used in 3D cell culture and highlight the unique benefits of using PeptiGel.