Exo-spin™ mini

Product description
Exo-spin™ products provide a proven, rapid and reliable way of generating high-quality purified exosomes suitable for a range of research applications.
Exo-spin™ technology combines precipitation and size exclusion chromatography (SEC), providing flexibility and performance. Using only precipitation for exosome isolation co-purifies large amounts of non-exosomal proteins and other material, as well as carried-over precipitant. SEC is reliable for exosome isolation, but a step is needed for low-exosome content starting material (such as cell culture media) to concentrate the sample prior to SEC isolation. For most applications, precipitation is the simplest way to achieve this concentration. Small samples can be purified directly. For larger samples we provide a simple, two-step protocol that allows consistent and reliable purification of samples in under 2 hours. Iterative loading can also be used to increase loading sample volume.
Exo-spin™ mini - maximum sample loading |
||||
|
Precipitant/concentration |
no |
yes |
no |
yes |
|
Iterative loading |
no |
no | yes |
yes |
|
Sera and plasma |
100 µl |
500 µl |
500 µl |
2500 µl |
|
culture media, urine, saliva |
100 µl |
50 ml |
500 µl |
250 ml |
Table 1. The maximum sample size that can be loaded onto a column will depend on the sample type and whether the sample is concentrated by precipitation or loaded iteratively.
Exo-spin™ column and resin pore size
The Exo-spin™ mini-column bed volume is 500 µl, allowing 100 µl of sample volume to be loaded on top of the column.
The resin used in these columns contains pores with a diameter of approximately 30 nm. All molecules which are smaller than 30 nm (e.g. free proteins, lipids) will enter into the pores and have their progress through the resin slowed. Using the protocol provided, these smaller particles remain trapped in the resin. Particles larger than 30nm are still small enough to fit between the resin beads and elute first. The first fraction to elute from the column contains particles between 30-200nm.
Highest recovery and purity
99% of proteins and lipids are removed by the column ensuring a highly pure sample ready for your downstream application.
Reproducibility
All our columns are manufactured in-house and undergo rigorous QC checks to ensure a high reproducibility between each lot. A large number of peer-reviewed scientific papers have been published describing the use of Exo-spin™.
Storage
Upon receipt, store purification Exo-spin™columns and Exo-spin™ Buffer should be stored at 4°C. All other components should be stored at room temperature (15°C - 25°C).
Frequently Asked Questions (FAQs)
For any additional questions, please refer to FAQs document below.
Product data
EX01 Exo-spin™ kit comparative data
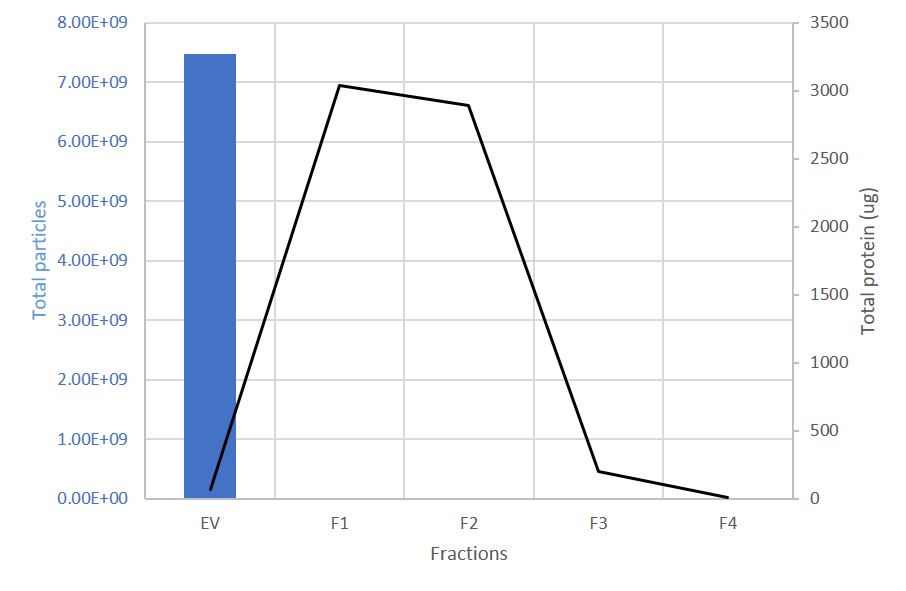
EV and protein content in fractions collected from columns. The first fraction (EV) containing the exosomes is retained. 99% of proteins are present in the subsequent, discarded fractions (F1-F4).
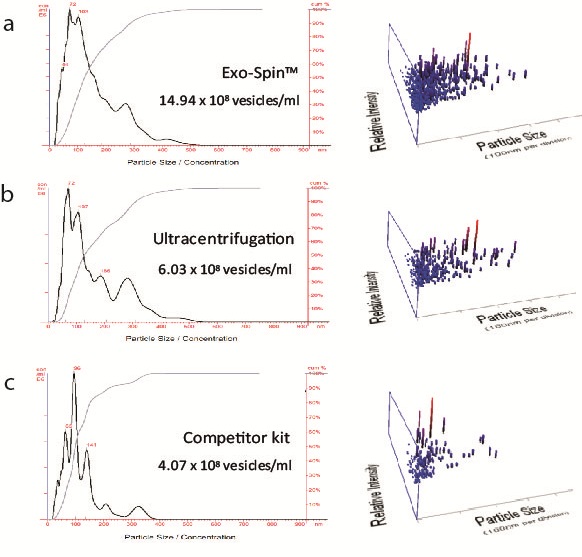
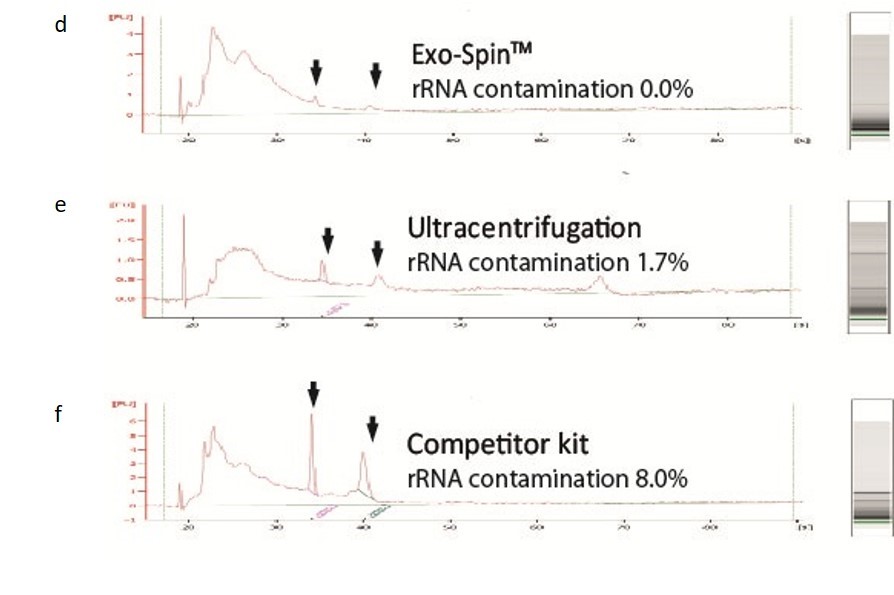
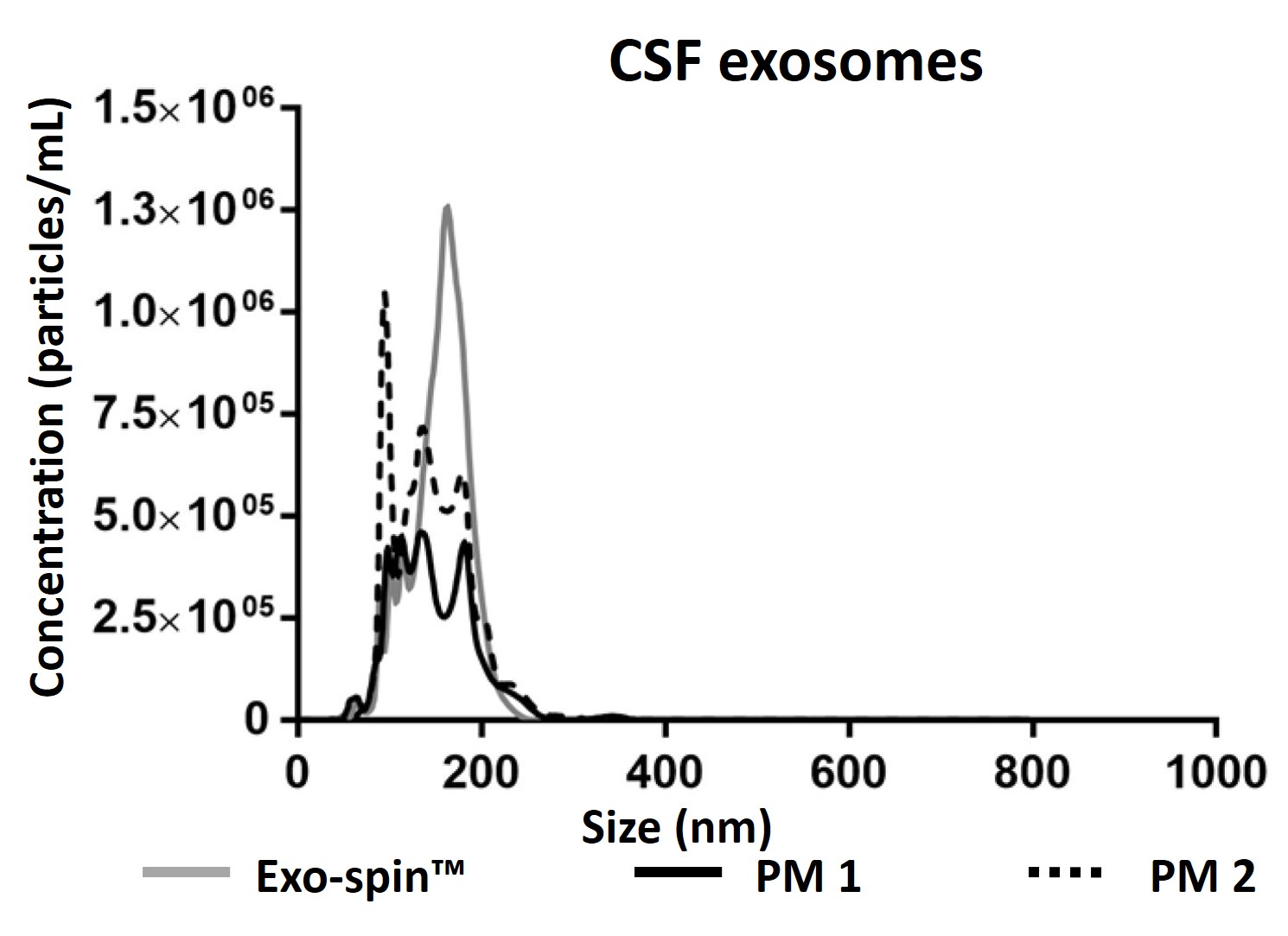
NTA measurement of samples prepared from cell culture medium: (a) Exo-spin™ yields significantly higher numbers of vesicles of the expected 40-120 nm size range than alternatives including (b) ultracentrifugation (c) competitor kit. The size distribution profile obtained with Exo-spin™ most closely resembles ultra-centrifugation. Lower rRNA contamination levels Analysis on Bioanalyzer instrument shows RNA preps (Trizol) following (d) Exo-spin™ purification from cell culture samples compared with (e) ultracentrifugation or (f) competitor kit.
Data determined by nanoparticle tracking analysis (NTA). Each curve represents the average of 3 technical replicate measurements for each exosome isolation method triplicate experiment. (PM = Precipitation Method). Figure taken and adapted from (Martins, TS et al., 2018).
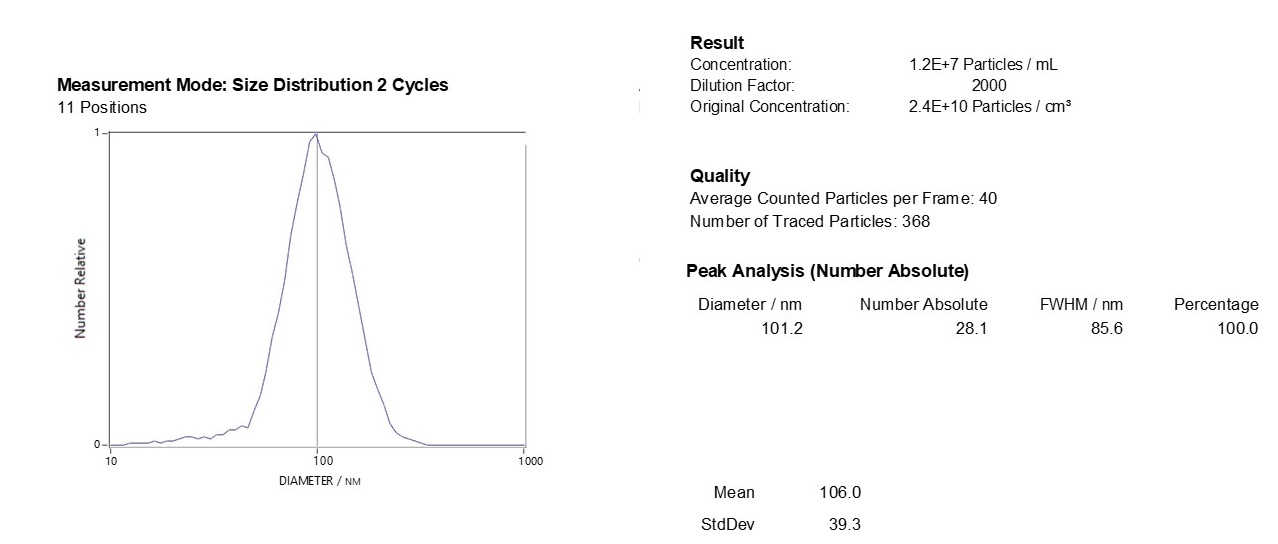
Characterizing Exo-spin™ isolated exosomes using nanoparticle tracking analysis (NTA)
Exosome characterization with NTA. Exosomes have been isolated using the Exo-spin™ kit and analysis performed with the ZetaView® instrument.
Downstream applications: advice and content
The EX01 Exo-spin™ kit is compatible with all downstream application and has been published in a large range of different applications.
RNA analysis:
- Li Z, Mbah NE and Maltese WA. (2018). Vacuole-inducing compounds that disrupt endolysosomal trafficking stimulate production of exosomes by glioblastoma cells. Molecular and Cellular Biochemistry 439(1-2):1-9.
- Koo KM, Wee EJ and Trau M.; (2016). Colorimetric TMPRSS2-ERG Gene Fusion Detection in Prostate Cancer Urinary Samples via Recombinase Polymerase Amplification. Theranostics 6(9): 1415–1424.
- Yoon C, Kim J, Park G, Kim S, Kim D, Hur DY, Kim B and Kim YS. (2016). Delivery of miR-155 to retinal pigment epithelial cells mediated by Burkitt's lymphoma exosomes. Tumor Biology 37(1):313-21.
Exosome engineering for therapeutic cargo:
- Kojima R, Bojar D, Rizzi G, Hamri GC, El-Baba MD, Saxena P, Ausländer S, Tan KR and Fussenegger M. (2018). Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nature communications 9: 1305.
Functional study:
- Piao YJ, Kim HS, Hwang EH, Woo J, Zhang M and Moon WK. (2017). Breast cancer cell-derived exosomes and macrophage polarization are associated with lymph node metastasis. Oncotarget 9(7): 7398–7410.
Proteomic (MS/MS analysis):
- E L Kavanagh, S Lindsay, M Halasz, L C Gubbins, K Weiner-Gorzel, M H Z Guang, A McGoldrick, E Collins, M Henry, A Blanco-Fernández, P O' Gorman, P Fitzpatrick, M J Higgins, P Dowling, and A McCann. (2017). Protein and chemotherapy profiling of extracellular vesicles harvested from therapeutic induced senescent triple negative breast cancer cells. Oncogenesis 6(10): e388.
Exosome labelling:
- Lara P, Palma-Florez S, Salas-Huenuleo E, Polakovicova I, Guerrero S, LobosGonzalez L, Campos A, Muñoz L, Jorquera-Cordero C, Varas-Godoy M, Cancino J, Arias E, Villegas J, Cruz LJ, Albericio F, Araya E, Corvalan AH, Quest AFG, Kogan MJ (2020). Gold nanoparticle based double-labeling of melanoma extracellular vesicles to determine the specificity of uptake by cells and preferential accumulation in small metastatic lung tumors. Journal of Nanobiotechnology 23;18(1):20.
Further resources:
Please see the Exosome Resources Page
Citations
There are more than 800 citations for Exo-spin products