Combining techniques improves extracellular vesicle isolate quality

Ultracentrifugation is often described as the “gold standard” to isolate or purify extracellular vesicles (EVs) including exosomes. Now, an EV study, led by researchers at Clemson University in South Carolina, has shown that for purification of EVs from urine samples, this gold standard can be improved with a simple modification.
EVs are of particular interest for diagnosing and monitoring disease. The researchers have a particular interest in monitoring EVs purified from urine to diagnose the early development of chronic kidney disease. If this disease’s development can be identified at an early stage, simple lifestyle adjustments may delay or prevent the onset of the disease which eventually requires kidney dialysis and transplant.
For EV disease marker identification, interpretation is simplified if contaminants (i.e non-EV material) are efficiently removed prior to analysis. The researchers decided to evaluate the yield and purity of samples purified using ultracentrifugation alone or ultracentrifugation followed by further clean-up using Exo-spin columns.
The study used urine samples collected from healthy volunteers. Each sample was subjected to either (A) ultracentrifugation or (B) ultracentrifugation plus Exo-spin preparation. Following purification, the samples were first subjected to NTA particle analysis. For each sample, the yields of EVs isolated in both preparation types was the same – the additional Exo-spin step did not reduce EV yield.
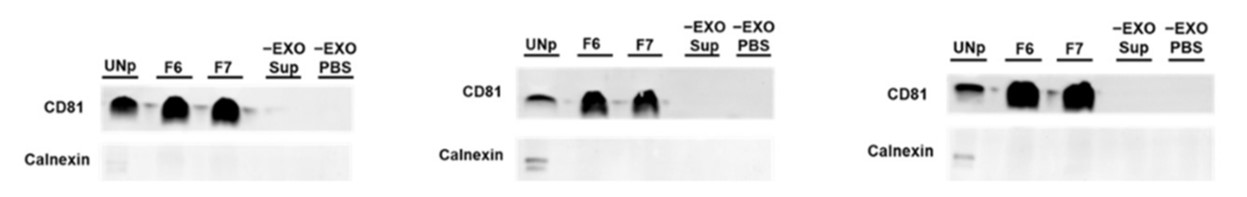
Next, immunoblotting of CD81, an exosome marker, and calnexin, an exosome negative marker was used to assess the purity of samples purified with (examples shown in F6 & F7 in the above image) and without Exo-spin (UNp). The intensity of the signal for samples that had been purified using Exo-spin was as high as (and in most cases higher than) the samples purified by ultracentrifugation alone. In contrast, the calnexin marker which revealed sample impurity was only visible in samples that had not benefitted from the additional Exo-spin step.
Finally, the researchers used transmission electron microscopy to confirm the presence of exosomes in the Exo-spin column purified samples.
The report concludes that for complex analysis (e.g. next-generation sequencing) of samples for marker discovery, the use of Exo-spin would facilitate analysis and interpretation of data by significantly reducing the level of non-EV contaminants. Exo-spin alone was not evaluated because the authors thought that it can't be used for large volume samples such as urine. However, precipitation of exosomes from these samples prior to Exo-spin purification using Exo-Spin buffer does enable efficient handling of these dilute exosome samples
The full manuscript for the study is available here. The figure used above is taken from the study.
MAIN IMAGE: SEM picture of kidney tubule. Credit: David Gregory & Debbie Marshall.