Karyotyping and chromosomal instability

Induced pluripotent stem cells (iPSCs) are cultured in many labs due to their potential in regenerative medicine, disease modelling and drug discovery. However, culture conditions and maintenance of these cells can lead to a loss of genetic integrity. These abnormalities are absent from donor cells, compromising their validity as disease models. Therefore, quality control is important to identify the presence of abnormalities or variants in chromosomes.
One of the key methods for identifying issues with chromosomes is karyotyping. This process involves culturing and harvesting cells to achieve metaphase chromosomes. Suitable cells are applied to slides and stained with a Giemsa stain that differentially stains areas rich in adenine and thymine DNA (darker staining) versus areas rich with guanine and cytosine (lighter staining) giving rise to the distinct and characteristic banding patterns called G-banding. After imaging, the chromosomes are then organised and arranged in numerical pair order (1 to 22 plus XY) to produce a karyogram for analysis.
Constitutional chromosome abnormalities are designated as present from birth and associated with embryonic cells. Chromosomal abnormalities that are seen, for example, in malignant tumours are referred to as “acquired” since they occur in differentiated somatic cells. They are hallmarks of gene dysregulation and genome instability. Human PSCs are usually genetically diploid but can acquire genetic abnormalities including chromosome changes, mutations and copy number variations (CNVs) with increased culture passage number. Using the karyotyping method these abnormalities are identified and defined in a number of ways.
Numerical chromosome abnormalities (also referred to as aneuploidy) are easily identified when chromosomes are organised by size and shape. Gains of an extra chromosome (trisomies) are found more often than losses (monosomies). Individual cells may contain multiple gains or losses. One of the most well-known, trisomy 21 (with an extra chromosome 21), is associated with Down’s syndrome. In iPSCs, this extra chromosome can confer a proliferative advantage, for example with trisomy 12 which is associated with the overexpression of the pluripotency gene NANOG. In other cases, the entire chromosomal set can be duplicated (polyploidy). Triploidy (69 chromosomes) and tetraploidy (96 chromosomes) are also known as euploidy.
As well as increases in chromosome number, iPSCs may also contain structural abnormalities. These abnormalities may not be detected by some newer technologies. When detecting gross structural rearrangements, the process of G-banding is particularly powerful. Changes seen can include:
- Deletion of chromosomal material.
- Translocation of material between two or more chromosomes.
- Duplication of chromosomal material.
- Inversion of chromosomal material
- Duplication of a chromosomal arm is referred to as an isochromosome.
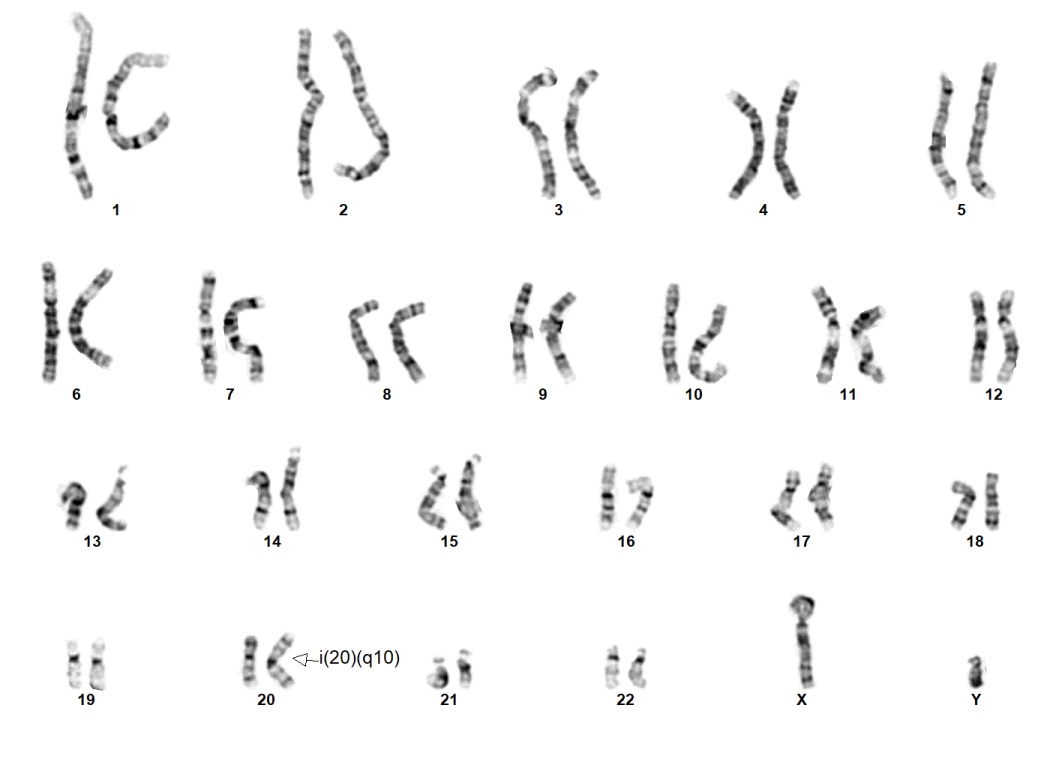
An abnormal iPSC karyotype 46,XY,i(20)(q10) This is the standard isochromosome 20.
Recurrent structural rearrangements can result from mechanisms for repairing broken chromosomes such as non-allelic homologous recombinations. Translocations and inversions often result in balanced chromosomal rearrangements. This type of abnormality does not manifest an apparent cytogenetic gain or loss of chromatin and so can be missed with newer high-resolution detection technologies. Thus, karyotyping remains the fundamental technique for detecting these abnormalities.
Mosaicism is almost always present in cell cultures at some level. Mosaicism is defined as the presence of genetic differences between two or more cells within a single analysed sample. Each of these cells is derived from the same original cell line. This can result in distinct structural and numerical abnormalities appearing in different cells within the same culture. The primary mechanisms leading to mosaicism formation include mitotic or meiotic non-disjunction flaws, anaphase lagging and trisomy rescue. Karyotyping is able to detect mosaicism in a single cell within a standard 20 iPSC cell analysis. Mosaicism is a potential concern when culturing hPSCs as it cannot reliably be detected at low levels (<10%) by many screening technologies leading to a negative impact on the research outcome. More research is needed to understand mosaicism, particularly due to its association with tumorigenic potential.
Abnormalities occur across the entire karyotype; however, aberrations are seen more commonly in respect to specific chromosomes with some of the most prevalent being chromosomes 1, 12 and 17. These are often a numerical gain and/or amplification on the p or q arm and are often recurrent. Losses on the q arm of chromosome 18 and the p arm of chromosome 10 occur less frequently but are still significant.
As with trisomy 12, there are associated candidate genes with these abnormalities which may explain the correlation with cell behaviour. For example, the gain seen on the q arm of chromosome 1 is related to the gene MDM4, a p53 suppressor, which increases the apoptosis threshold. Additionally, an identified driver gene with the gains seen in chromosome 17 is the anti-apoptotic gene BIRC5. Both of these abnormalities offer a proliferative advantage to cells and therefore drive selection pressure towards these types of aberrant cells. Studies indicate that 20% of cells carry at least one megabase of chromosomal anomaly, and this may even be an underestimation due to current detection limitations.
IMAGE: Cell Guidance Systems
Learn more about powerful technologies that are enabling research:
Recent technological advances have provided faster methods with higher resolution. However, they may not detect all types of abnormality or provide readily interpretable data. Karyotyping remains the gold standard as an effective, low-cost method that can identify balanced and unbalanced chromosomal rearrangements, polyploidy, and mosaicism. As an adjunct to karyotyping, SNP-arrays allow analysis to a 200kb resolution, compared to approximately 10Mb offered by karyotyping. Although it still has inherent limitations, such as inability to detect balanced translocations, arrays' ability to detect smaller genomic imbalances make it a useful complementary technique to karyotyping.